Katie Lotterhos FSU Department of Biological Sciences, FSU
When we look at a salt marsh, we see thousands of stems of cordgrass. But in reality, the coastline may be made up of only a few different genetic individuals. This is because Spartina can spread by growing clones of itself, with the exact same genetic code (a genotype). Why does it matter if we know whether or not a salt marsh is made up of one or many different genotypes? Well, different genotypes will have different abilities to resist pests or disease, or they may be tastier to eat for the little marsh critters like snails and grasshoppers. Since some genotypes will be better than others in different situations, we care about genetic diversity because it can be a buffer against an uncertain environment.
Since we can’t just look at a marsh and tell which individuals have which genotypes, that’s where genetics come in. In each individual, we analyze short pieces of the genetic code called microsatellites that all individuals have, but that vary in length in different individuals. If individuals have the same microsatellite lengths or genetic “fingerprint”, then they are part of the same clone.
It’s not magic, and this is how we do it….
<———This is a case of liquid nitrogen. We clip a piece of plant tissue from each individual, label the location, and put it on ice.
——————————————> This is a warm water bath. We digest the plant tissue, leaving only the DNA and other molecules.
<————These are magnetic beads. Since DNA is polarized (it has a positively-charged side and a negatively-charged side), it sticks to the beads. This purifies the DNA.
———————————————->This is the fluorescently-tagged forward primer for the microsatellite. We use these primers to amplify only the part of the DNA that we are interested in. We need many copies of DNA to get a fluorescent signal. We mix this with a reverse primer and the DNA of each individual in a tube with buffer.
 <——————This is a PCR machine. It heats up the DNA, which causes it to split apart. Then it cools the DNA, and then the primers bind to the DNA and with the special buffer, it makes a copy of that region of the genome (the microsatellite!). It then heats and cools several times to make thousands of copies of the small region we are interested in.
<——————This is a PCR machine. It heats up the DNA, which causes it to split apart. Then it cools the DNA, and then the primers bind to the DNA and with the special buffer, it makes a copy of that region of the genome (the microsatellite!). It then heats and cools several times to make thousands of copies of the small region we are interested in.
————————-> This is the good luck charm that I keep by the PCR machine. Sometimes getting PCRs to work involves praying, voodoo, sacrifices, swearing, headstands, cartwheels, magic, or just plain luck (or all of the above).
<————–This is the sequencing machine that reads the fluorescent signal. After PCR, we send the product to this machine and it gives us the output.
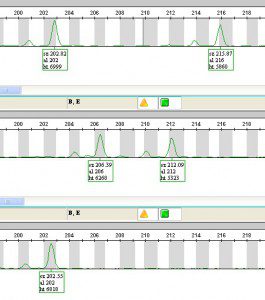 ———————->This is the fluorescent signal, showing the length of the microsatellite! Each row in this picture shows a different individual, and their florescent peaks (each one can have 2 peaks, one from each chromosome). These individuals all have different genotypes (or microsatellite lengths). Each clone (which could be several individuals) has its own unique fluorescent “fingerprint.”
———————->This is the fluorescent signal, showing the length of the microsatellite! Each row in this picture shows a different individual, and their florescent peaks (each one can have 2 peaks, one from each chromosome). These individuals all have different genotypes (or microsatellite lengths). Each clone (which could be several individuals) has its own unique fluorescent “fingerprint.”
.
.
So that’s it! And that’s how we can take a marsh that looks like this:
 and turn it into a map like this:
and turn it into a map like this:
 in which the different colors represent different clones in the population.
in which the different colors represent different clones in the population.









2 comments
[…] read about Emily Field’s graduate work on seagrass wrack and Kattie Lotterhos’ graduate work in genetics. In David’s lab, we’ve heard from Tanya Rogers, a lab technician who keeps […]
[…] at a variety of cordgrass individuals, or just one. You wouldn’t know until you got the DNA results back from the lab. That’s genetic diversity, the variation of genes within a species. A little more obvious […]
Comments are closed.